metan v1.6.1 on CRAN
I’m so excited to announce that the latest stable version (v1.6.1) of the R package metan is now on
CRAN. The main features included in this version are detailed below.
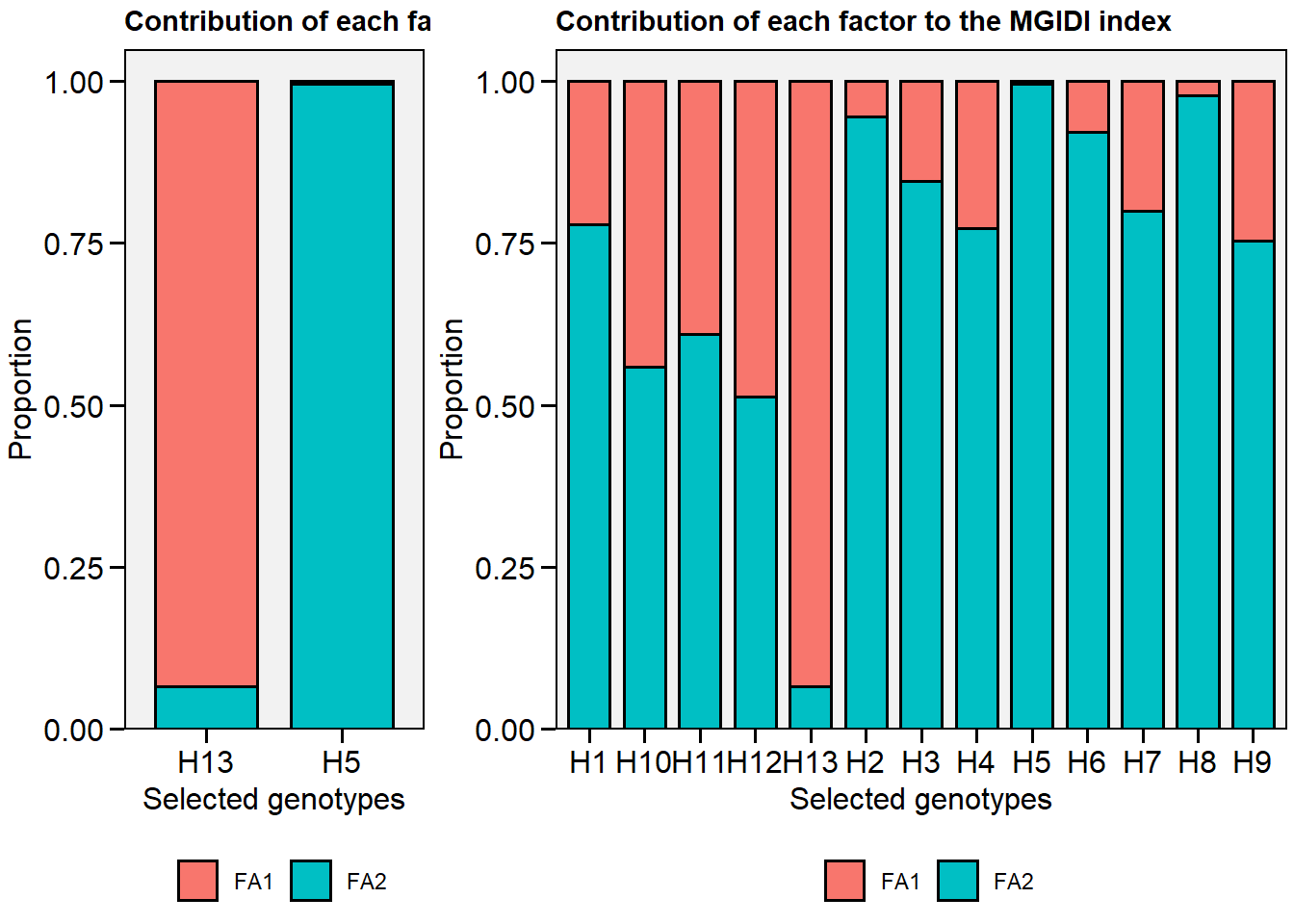
plot.mgidi()can now plot the contribution for all genotypes.
In metan v1.6.1, we can plot the contribution of the factors to the MGIDI index for all genotypes, including the arguments type = "contribution" and genotypes == "all"
library(metan)
# Registered S3 method overwritten by 'GGally':
# method from
# +.gg ggplot2
# []=====================================================[]
# [] Multi-Environment Trial Analysis (metan) v1.6.1 []
# [] Author: Tiago Olivoto []
# [] Type citation('metan') to know how to cite metan []
# [] Type vignette('metan_start') for a short tutorial []
# [] Visit http://bit.ly/2TIq6JE for a complete tutorial []
# []=====================================================[]
model <- gamem(data_g,
gen = GEN,
rep = REP,
resp = c(NR, KW, CW, CL, NKE, TKW, PERK, PH))
# Method: REML/BLUP
# Random effects: GEN
# Fixed effects: REP
# Denominador DF: Satterthwaite's method
# ---------------------------------------------------------------------------
# P-values for Likelihood Ratio Test of the analyzed traits
# ---------------------------------------------------------------------------
# model NR KW CW CL NKE TKW PERK PH
# Complete NA NA NA NA NA NA NA NA
# Genotype 0.0056 0.0253 1.24e-05 2.25e-06 0.00952 0.00955 4.65e-07 0.051
# ---------------------------------------------------------------------------
# Variables with nonsignificant Genotype effect
# PH
# ---------------------------------------------------------------------------
# Selection for increase all variables
mgidi_model <- mgidi(model)
#
# -------------------------------------------------------------------------------
# Principal Component Analysis
# -------------------------------------------------------------------------------
# # A tibble: 8 x 4
# PC Eigenvalues `Variance (%)` `Cum. variance (%)`
# <chr> <dbl> <dbl> <dbl>
# 1 PC1 3.89 48.6 48.6
# 2 PC2 3.09 38.6 87.1
# 3 PC3 0.518 6.48 93.6
# 4 PC4 0.272 3.40 97.0
# 5 PC5 0.178 2.23 99.2
# 6 PC6 0.0561 0.702 100.
# 7 PC7 0.00302 0.0377 100.
# 8 PC8 0.000966 0.0121 100
# -------------------------------------------------------------------------------
# Factor Analysis - factorial loadings after rotation-
# -------------------------------------------------------------------------------
# # A tibble: 8 x 5
# VAR FA1 FA2 Communality Uniquenesses
# <chr> <dbl> <dbl> <dbl> <dbl>
# 1 NR -0.0653 -0.899 0.813 0.187
# 2 KW -0.444 -0.842 0.907 0.0930
# 3 CW -0.935 -0.280 0.953 0.0468
# 4 CL -0.952 0.0294 0.907 0.0926
# 5 NKE 0.405 -0.853 0.891 0.109
# 6 TKW -0.913 -0.000960 0.834 0.166
# 7 PERK 0.933 -0.154 0.894 0.106
# 8 PH 0.0195 -0.879 0.772 0.228
# -------------------------------------------------------------------------------
# Comunalit Mean: 0.8713971
# -------------------------------------------------------------------------------
# Selection differential
# -------------------------------------------------------------------------------
# # A tibble: 8 x 10
# VAR Factor Xo Xs SD SDperc h2 SG SGperc sense
# <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr>
# 1 CW FA 1 20.8 23.7 2.95 14.2 0.880 2.60 12.5 increase
# 2 CL FA 1 28.4 28.7 0.300 1.06 0.901 0.270 0.950 increase
# 3 TKW FA 1 318. 314. -3.67 -1.16 0.712 -2.62 -0.824 increase
# 4 PERK FA 1 87.6 87.6 -0.0589 -0.0672 0.916 -0.0539 -0.0615 increase
# 5 NR FA 2 15.8 17.4 1.63 10.3 0.736 1.20 7.60 increase
# 6 KW FA 2 147. 163. 16.2 11.0 0.659 10.7 7.27 increase
# 7 NKE FA 2 468. 532. 64.0 13.7 0.713 45.6 9.74 increase
# 8 PH FA 2 2.17 2.28 0.110 5.07 0.610 0.0670 3.09 increase
# ------------------------------------------------------------------------------
# Selected genotypes
# -------------------------------------------------------------------------------
# H13 H5
# -------------------------------------------------------------------------------
# plot the contribution of each factor on the MGIDI index
p1 <- plot(mgidi_model, type = "contribution")
p2 <- plot(mgidi_model, type = "contribution", genotypes = "all")
arrange_ggplot(p1, p2, rel_widths = c(1, 2))
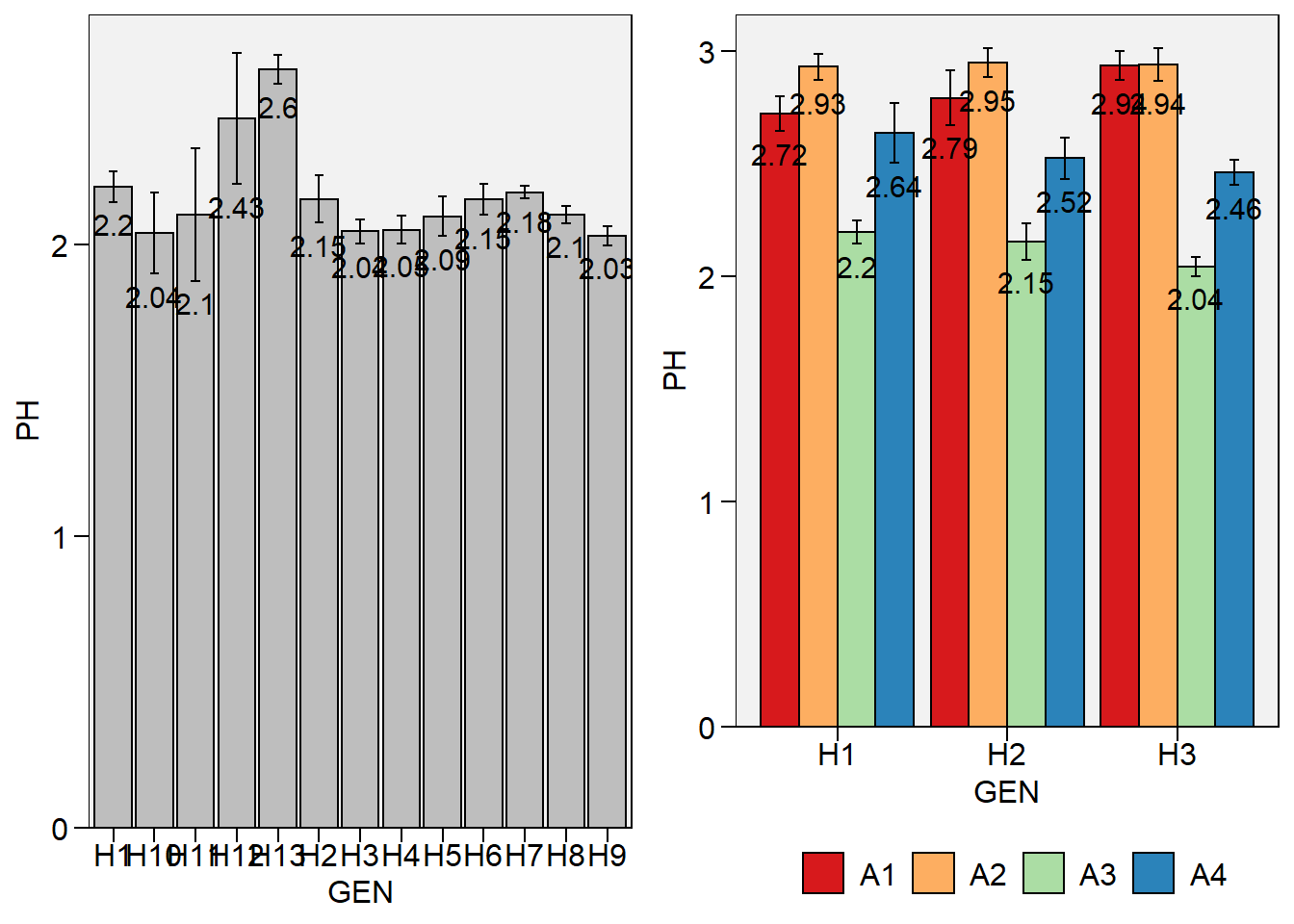
plot_bars()andplot_factbars()now shows the values withvalues = TRUE
# one categorical variable
p1 <- plot_bars(data_g, GEN, PH, values = TRUE)
p2 <-
data_ge2 %>%
subset(GEN %in% c("H1", "H2", "H3")) %>%
plot_factbars(GEN, ENV, resp = PH, values = TRUE)
arrange_ggplot(p1, p2)
# Warning: Graphs cannot be horizontally aligned unless the axis parameter is set.
# Placing graphs unaligned.
- Update the functions by using the new
dplyr::across() - Update citation field by including number and version of the published paper.
citation("metan")
#
# Please, support this project by citing it in your publications!
#
# Olivoto, T., and Lúcio, A.D. (2020). metan: an R package for
# multi-environment trial analysis. Methods Ecol Evol. 11:783-789
# doi:10.1111/2041-210X.13384
#
# A BibTeX entry for LaTeX users is
#
# @Article{Olivoto2020,
# author = {Tiago Olivoto and Alessandro Dal'Col L{'{u}}cio},
# title = {metan: an R package for multi-environment trial analysis},
# journal = {Methods in Ecology and Evolution},
# volume = {11},
# number = {6},
# pages = {783-789},
# year = {2020},
# doi = {10.1111/2041-210X.13384},
# url = {https://besjournals.onlinelibrary.wiley.com/doi/abs/10.1111/2041-210X.13384},
# eprint = {https://besjournals.onlinelibrary.wiley.com/doi/pdf/10.1111/2041-210X.13384},
# }
Follow metan on Github to find out the latest news!