Production progress
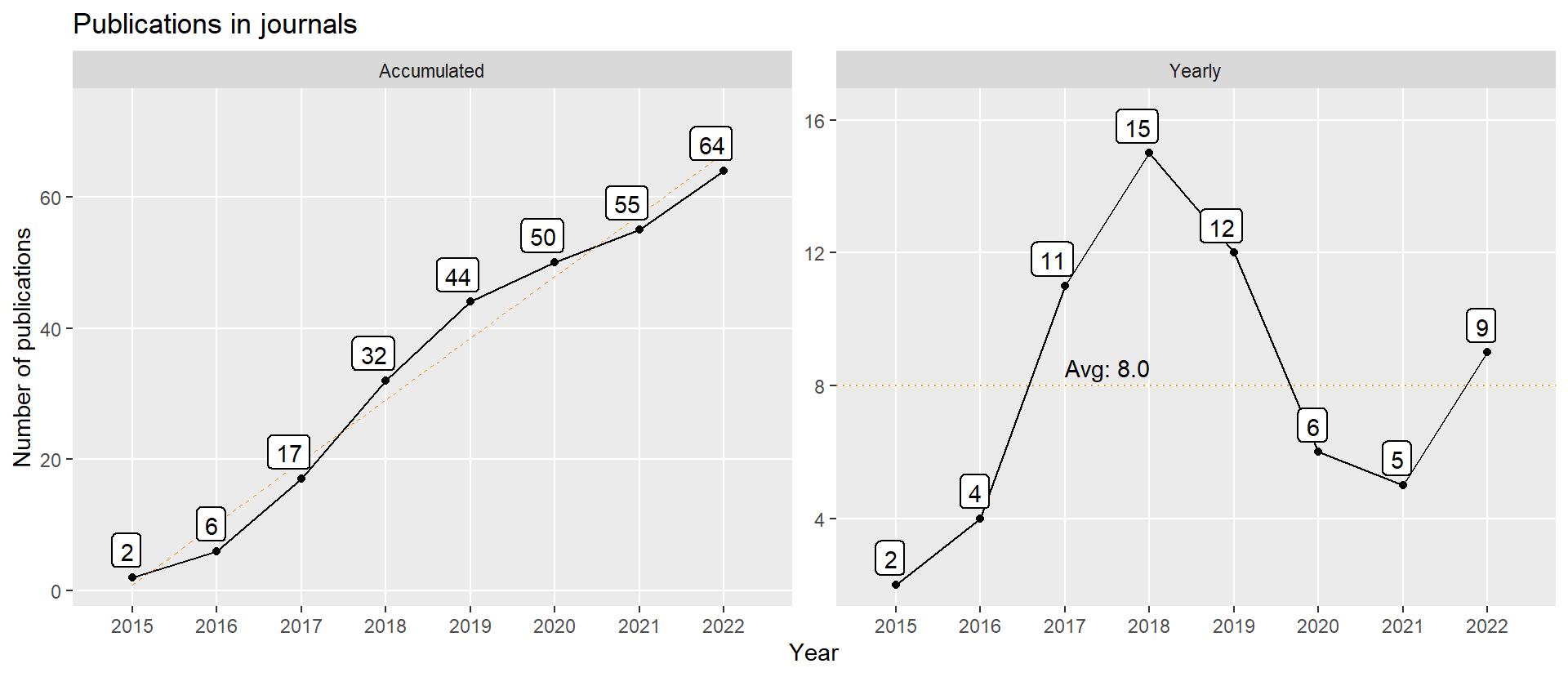
Figure 1: Number of publications in journal over time. The complete list of references is at the bottom of the page. Last update at 2022-12-27.
Citation progress
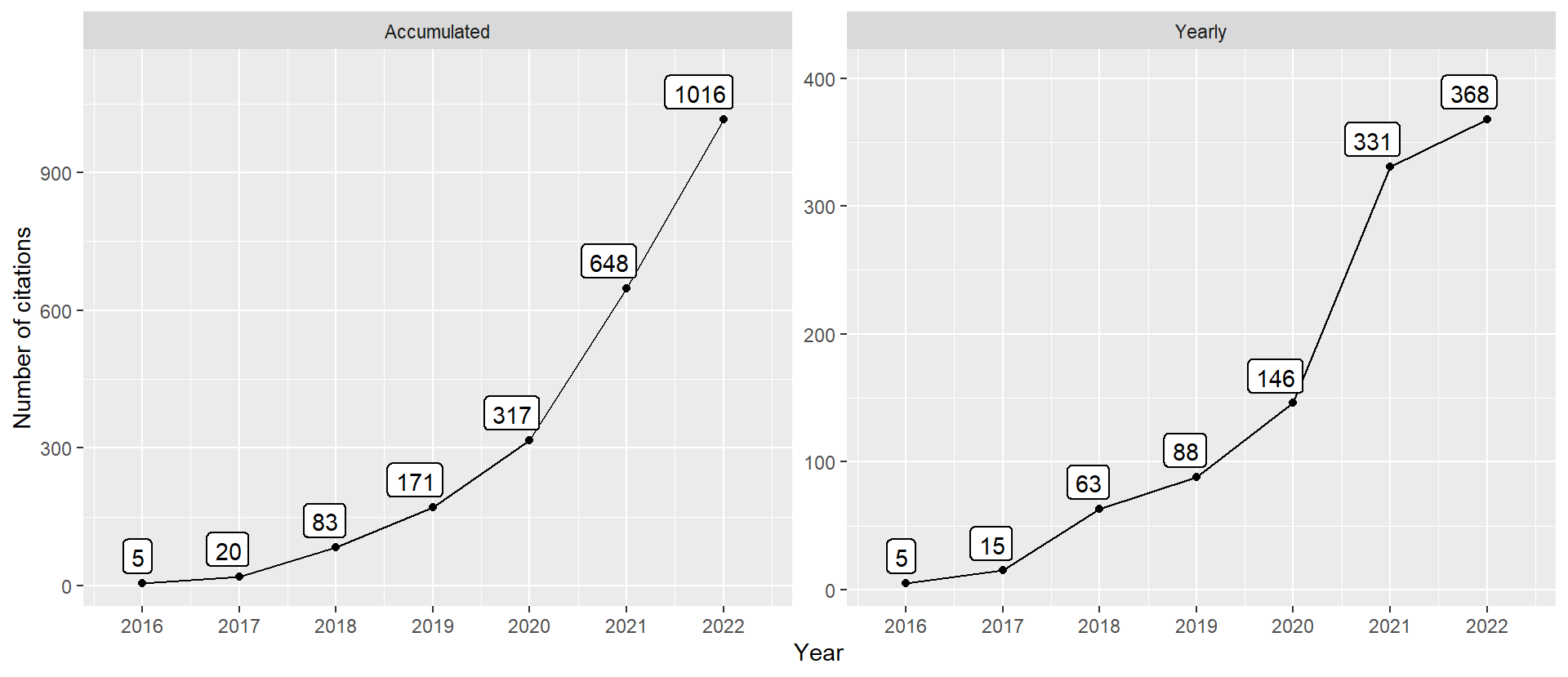
Figure 2: Number of citations over time. Last update at 2022-12-27.
Doctor Thesis
Genotypic stability indexes and multivariate simultaneous selection: a new approach
Abstract: In order to better understand and explore the genotype-environment interaction (GEI) in plant breeding, the development of new methods for adaptability and stability analysis, as well as the improvement of existing ones, is necessary. This study introduces the theoretical foundations, shows the numerical application and the implementation into a statistical software of new indexes for genotypic stability and multivariate simultaneous selection in plant breeding. The singular value decomposition of a two-way matrix containing the BLUPs (Best Linear Unbiased Prediction) of the GEI effects obtained in a linear mixed-effect model (LMM) was used to produce biplots useful in identifying the patterns of a random structure of GEI. A new quantitative index of genotypic stability called WAASB, based on the weighted average of the absolute value decomposition scores of the BLUPs matrix for the effects of IGA obtained in an MLM is proposed. By definition, the lower the WAASB value, the more stable a given genotype is. It is also introduced the theoretical foundations of a superiority index that allows weighting between stability (WAASB) and mean performance (Y), which was conveniently called WAASBY. The WAASBY assumes values in the range of 0−100, with 100 being assigned to the ideotype, i.e., the genotype that was most stable and that best performed on average among those considered in the test environments. A multi-trait stability index (MTSI) is used to extend the WAASB and WAASBY indexes to a multivariate structure, thus allowing selection for stability or simultaneous selection for stability and mean performance based on several traits. The application of these indexes is illustrated using real data from multi-environment trials with white oat (Avena sativa L.) crop. The WAASB allowed the quantification of genotypic stability and the identification of genotype groups with different patterns for stability and mean performance. Using the WAASBY index it was possible to identify genotypes that combine simultaneously high performance and yield stability. In the context of multivariate selection, positive selection differentials (SD) (1.75% ≤ SD ≤ 17.8%) were observed for trait means that wanted to increase and negative (SD = −11.7%) for one variable that wanted to reduce. The negative DS obtained for the WAASB index (−63% ≤ SD ≤ −12%) suggesting that the selected genotypes were more stable. Reliable stability measures using WAASB can help breeders and agronomists make the right decisions when selecting or recommending genotypes. Besides, the simultaneous selection index, WAASBY, will be useful when selection considers different weights for stability and mean performance. The MTSI has broad applicability in simultaneous selection for stability and mean performance based on multiple traits since it provides a unique selection process that is easy-to-handle and considers the correlation structure between traits. The proposed indices were implemented in the R metan (multi-environment trial analysis) software package. The development version of metan is available on Github https://tiagoolivoto.github.io/metan/ and can be installed directly via console R using devtools::install_github("TiagoOlivoto/metan"). The package metan presents a collection of functions for verifying, manipulating and summarizing typical multi-environment trial data, analyzing single-environment trials using both fixed- and mixed-effect models, computing parametric and non-parametric stability statistics, and implementing multivariate analysis.
Keywords: AMMI. BLUP. Genotype−environment interaction. GGE. metan.
Committee (Approved on December 19, 2019):
- Prof. Dr. Alessandro Dal’Col Lúcio (Orientador) - UFSM
- Prof. Dr. José Antonio Gonzales da Silva - UNIJUÍ
- Prof. Dr. Sidinei José Lopes - UFSM
- Prof. Dr. Thomas Newton Martin - UFSM
- Prof. Dr. Velci Queiróz de Souza - UNIPAMPA
Master Thesis
Bias associated with data arrangement and sample size and its implications on the accuracy of indirect selection in plant breeding
Abstract: Some data arrangement methods currently used may overestimate Pearson correlation coefficient (r) among explanatory traits, increasing multicollinearity in analysis that uses multiple regression. In this sense, the aims of the present research were to reveal the impact of different data arrangement scenarios on the multicollinearity of matrices, on the efficiency of the used methods to adjust it, on the estimates of coefficients and accuracy of the path analysis, as well as to use simulations to reveal the statistical behavior of the r and the optimal sample size for estimating r between maize traits. For this, data from an experiment conducted in a randomized complete design in a 15 × 3 factorial scheme (15 maize hybrids × three growing sites), arranged in four replicates were used. The traits analyzed in five plants of each plot were: plant height, ear insertion height, diameter and length of ear, number of rows per ear, number of kernels per row, diameter and length of cob, cob diameter/ear diameter ratio, number of kernels per ear, kernel mass per ear and thousand-kernel weight. At first, three path analysis methods (traditional, with k inclusion and with the exclusion of traits) having as a dependent trait the kernel mass per ear were tested in two scenarios: 1) with the linear correlation matrix (X’X) between the traits estimated with all sampled observations, n = 900 and 2) with the X’X matrix estimated with the average value of the five sampled plants in each plot, n = 180. Subsequently, aiming to evaluate the statistical behavior of r, in addition to the two described scenarios, the average value of treatments at each site, n = 45, was also considered. In each scenario, 60 sample sizes were simulated by using bootstrap simulations with replacement. Confidence intervals for combinations of different magnitudes were estimated in each scenario and sample size. One hundred and eighty correlation matrices (three scenarios × 60 sample sizes) were estimated and the multicollinearity evaluated. The number of kernels per ear and the thousand-kernel weight presented the most expressive direct effects to kernel mass per ear (r = 0.892 and r = 0.733, respectively). The use of average values reduces the individual variance of a set of n-traits, overestimates the magnitude of the r between the trait pairs, increases the multicollinearity of the matrix, and reduces the effectiveness of the used methods to adjust it as well as the accuracy of the path coefficient estimates. The number of plants required to estimate correlation coefficients with a 95% bootstrap confidence interval is greater when all sampled observations are used and increases in the sense of combination pairs with lower magnitude. By using all sampled observations, 210 plants are sufficient to estimate r between traits of simple maize hybrids in the 95% bootstrap confidence interval < 0.30. A simple method that reduces the multicollinearity of matrices and improves the accuracy of path analysis is proposed.
Keywords: Zea mays L. Correlation coefficient. Multicollinearity. Simulations.
Committee (Approved on February 20, 2017):
- Prof. Dr. Velci Queiróz de Souza (Orientador) - UFSM
- Prof. Dr. Volmir Sério Marchioro - UFSM
- Prof. Dr. Marcos Vinícios Marques Pinheiro - UFSM
Complete CV
Please, visit my profile at any of the services below:

