Inspeção de dados
Conjunto de dados
library(metan)
library(rio)
# gerar tabelas html
print_tbl <- function(table, digits = 3, ...){
knitr::kable(table, booktabs = TRUE, digits = digits, ...)
}
# Dados "arrumados"
df_tidy <- import("http://bit.ly/df_tidy", setclass = "tbl")
Inspecionar dados
inspect(df_tidy)
## # A tibble: 13 x 9
## Variable Class Missing Levels Valid_n Min Median Max Outlier
## <chr> <chr> <chr> <chr> <int> <dbl> <dbl> <dbl> <dbl>
## 1 ENV character No 0 114 NA NA NA NA
## 2 GEN character No 0 114 NA NA NA NA
## 3 BLOCO character No 0 114 NA NA NA NA
## 4 ALT_PLANT numeric No - 114 0 2.52 3.04 1
## 5 ALT_ESP numeric No - 114 0.75 1.34 1.88 0
## 6 COMPES numeric No - 114 12.2 15.1 17.9 0
## 7 DIAMES numeric No - 114 43.5 49.8 54.9 0
## 8 COMP_SAB numeric No - 114 23.5 28.2 34.7 0
## 9 DIAM_SAB numeric No - 114 13.3 16.0 18.3 0
## 10 MGE numeric No - 114 106. 172. 251. 0
## 11 NFIL numeric No - 114 12.4 16 21.2 1
## 12 MMG character No 0 114 NA NA NA NA
## 13 NGE numeric Yes - 112 354 504. 5302 5
## Warning: Considering the levels of factors, .data should have 1 rows, but it has
## 114. Use 'as_factor()' for coercing a variable to a factor.
## Warning: Expected three or more factor variables. The data has only 0.
## Warning: Missing values in variable(s) NGE.
## Warning: Possible outliers in variable(s) ALT_PLANT, NFIL, NGE. Use
## 'find_outliers()' for more details.
## Warning: Zero values observed in variable(s) ALT_PLANT.
# converter as três primeiras colunas para fator
df_ok <-
df_tidy %>%
metan::as_factor(1:3)
Fragmentos de texto inesperados
# Encontrar fragmentos de texto
find_text_in_num(df_ok, MMG)
## [1] 112
df_ok[112, 12]
## # A tibble: 1 x 1
## MMG
## <chr>
## 1 335..2
# substitui '..' por '.' e converte para numérico
df_ok <-
df_ok %>%
replace_string(MMG, pattern = "\\.{2}", replacement = ".") %>%
as_numeric(MMG)
Substituir zeros por NA
df_ok <-
df_ok %>%
replace_zero(ALT_PLANT)
# Nova inspeção
inspect(df_ok, plot = TRUE)
## # A tibble: 13 x 9
## Variable Class Missing Levels Valid_n Min Median Max Outlier
## <chr> <chr> <chr> <chr> <int> <dbl> <dbl> <dbl> <dbl>
## 1 ENV factor No 3 114 NA NA NA NA
## 2 GEN factor No 13 114 NA NA NA NA
## 3 BLOCO factor No 3 114 NA NA NA NA
## 4 ALT_PLANT numeric Yes - 113 1.71 2.52 3.04 0
## 5 ALT_ESP numeric No - 114 0.75 1.34 1.88 0
## 6 COMPES numeric No - 114 12.2 15.1 17.9 0
## 7 DIAMES numeric No - 114 43.5 49.8 54.9 0
## 8 COMP_SAB numeric No - 114 23.5 28.2 34.7 0
## 9 DIAM_SAB numeric No - 114 13.3 16.0 18.3 0
## 10 MGE numeric No - 114 106. 172. 251. 0
## 11 NFIL numeric No - 114 12.4 16 21.2 1
## 12 MMG numeric No - 114 227. 336. 452. 0
## 13 NGE numeric Yes - 112 354 504. 5302 5
## Warning: Considering the levels of factors, .data should have 117 rows, but it
## has 114. Use 'as_factor()' for coercing a variable to a factor.
## Warning: Missing values in variable(s) ALT_PLANT, NGE.
## Warning: Possible outliers in variable(s) NFIL, NGE. Use 'find_outliers()' for
## more details.

Outliers
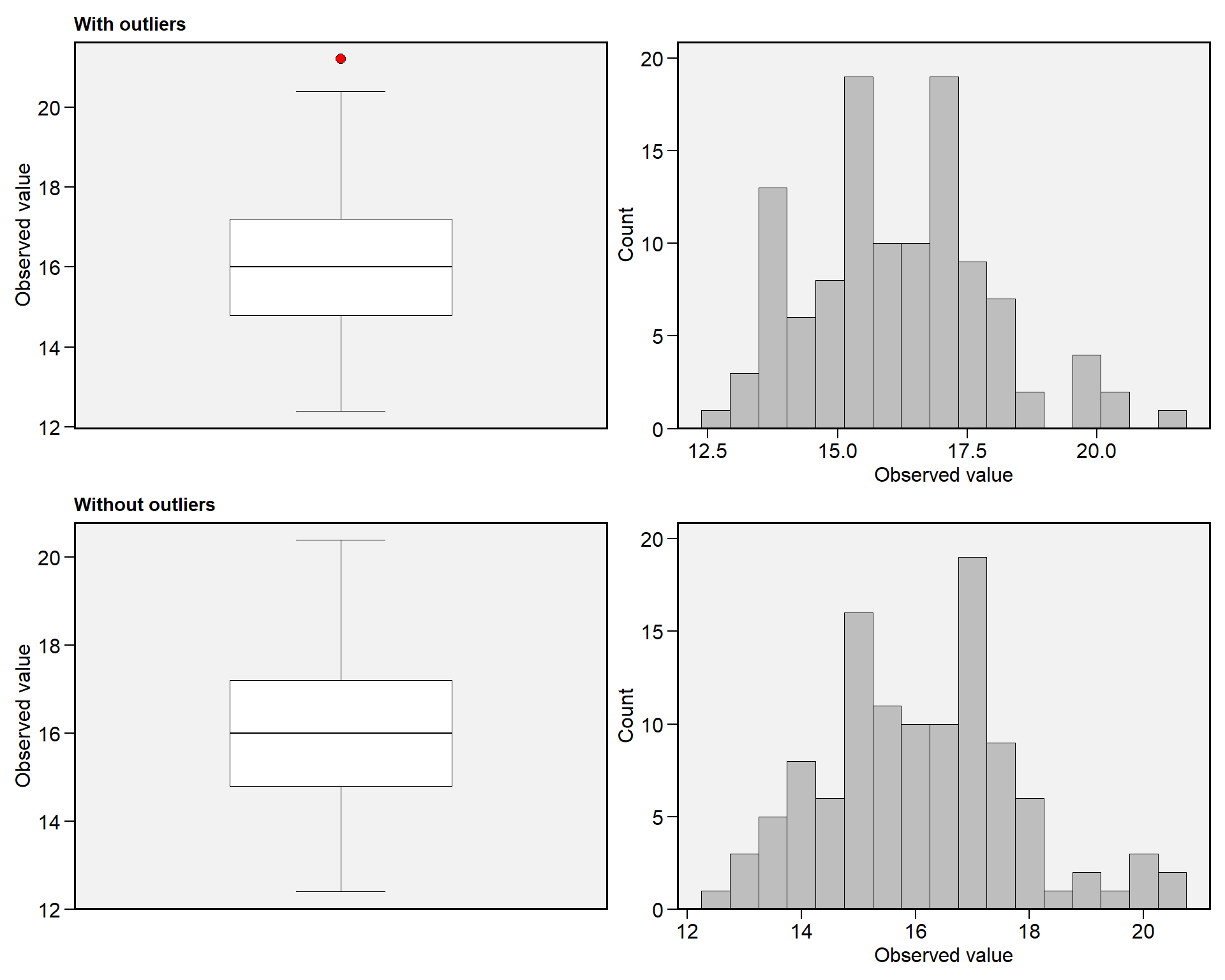
# Outlier NFIL
find_outliers(df_ok, NFIL, plots = TRUE)
## Trait: NFIL
## Number of possible outliers: 1
## Line(s): 14
## Proportion: 0.9%
## Mean of the outliers: 21.2
## Maximum of the outliers: 21.2 | Line 14
## Minimum of the outliers: 21.2 | Line 14
## With outliers: mean = 16.154 | CV = 10.833%
## Without outliers: mean = 16.11 | CV = 10.499%
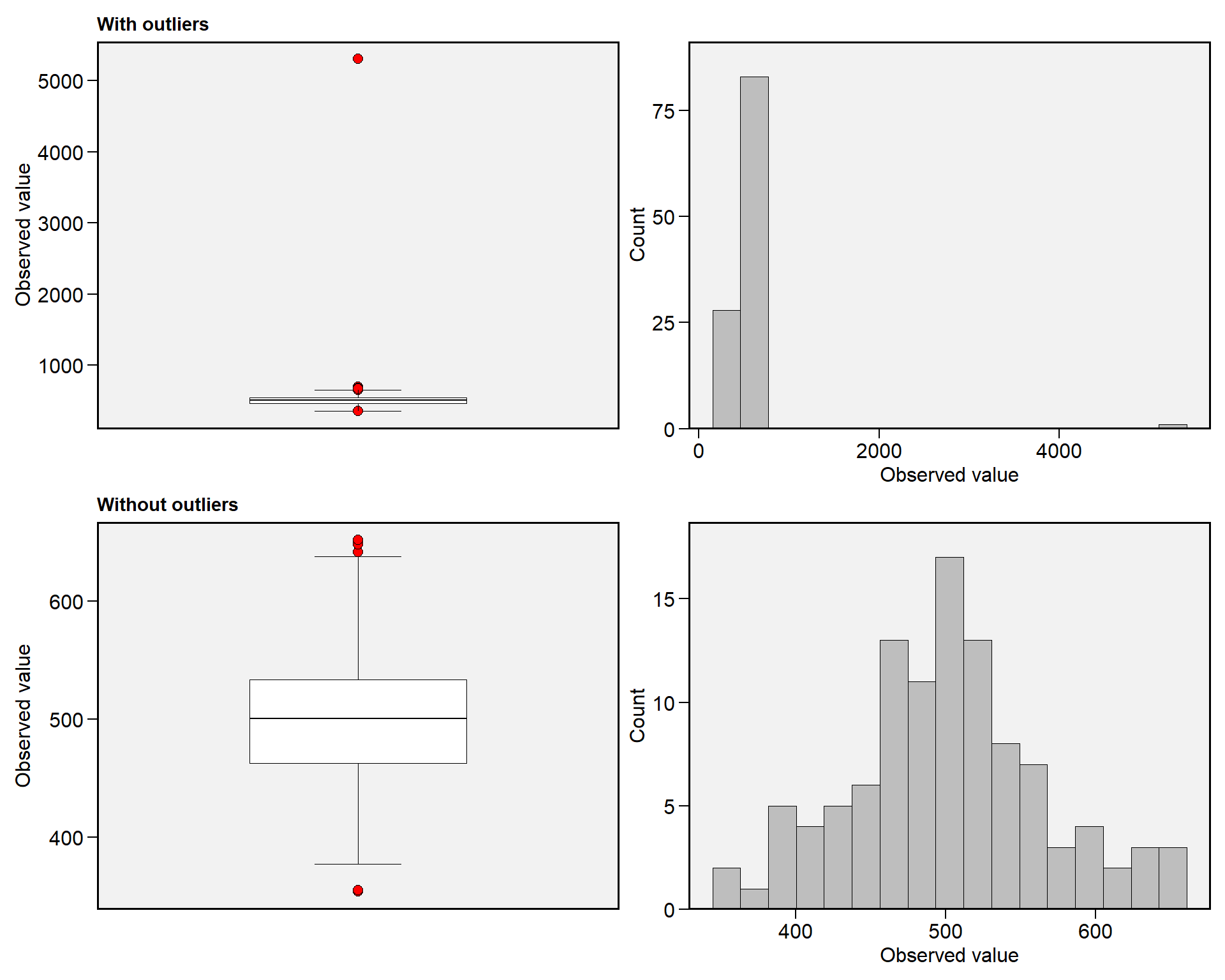
# Outlier NGE
find_outliers(df_ok, NGE, plots = TRUE)
## Trait: NGE
## Number of possible outliers: 5
## Line(s): 11 14 22 66 89
## Proportion: 4.7%
## Mean of the outliers: 1606.8
## Maximum of the outliers: 5302 | Line 22
## Minimum of the outliers: 667.8 | Line 89
## With outliers: mean = 550.623 | CV = 83.324%
## Without outliers: mean = 501.269 | CV = 13.121%
# Corrigir valores
df_ok[22, 13] <- 530.2
# Exportar df_ok
# export(df_ok, "df_ok.xlsx")