library(metan)
library(rio)
library(emmeans)
# gerar tabelas html
print_tbl <- function(table, digits = 3, ...){
knitr::kable(table, booktabs = TRUE, digits = digits, ...)
}
# dados
df <- import("http://bit.ly/df_ge", setclass = "tbl")
print(df)
## # A tibble: 156 x 13
## ENV GEN BLOCO ALT_PLANT ALT_ESP COMPES DIAMES COMP_SAB DIAM_SAB MGE
## <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 A1 H1 I 2.61 1.71 16.1 52.2 28.1 16.3 217.
## 2 A1 H1 II 2.87 1.76 14.2 50.3 27.6 14.5 184.
## 3 A1 H1 III 2.68 1.58 16.0 50.7 28.4 16.4 208.
## 4 A1 H10 I 2.83 1.64 16.7 54.1 31.7 17.4 194.
## 5 A1 H10 II 2.79 1.71 14.9 52.7 32.0 15.5 176.
## 6 A1 H10 III 2.72 1.51 16.7 52.7 30.4 17.5 207.
## 7 A1 H11 I 2.75 1.51 17.4 51.7 30.6 18.0 217.
## 8 A1 H11 II 2.72 1.56 16.7 47.2 28.7 17.2 181.
## 9 A1 H11 III 2.77 1.67 15.8 47.9 27.6 16.4 166.
## 10 A1 H12 I 2.73 1.54 14.9 47.5 28.2 15.5 161.
## # ... with 146 more rows, and 3 more variables: NFIL <dbl>, MMG <dbl>,
## # NGE <dbl>
Anova individual - anova_ind()
ind_an <- anova_ind(df,
env = ENV,
gen = GEN,
rep = BLOCO,
resp = everything(),
verbose = FALSE)
print(ind_an)
## Variable ALT_PLANT
## ---------------------------------------------------------------------------
## Within-environment ANOVA results
## ---------------------------------------------------------------------------
## # A tibble: 4 x 15
## ENV MEAN DFG MSG FCG PFG DFB MSB FCB PFB DFE
## <chr> <dbl> <int> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <int>
## 1 A1 2.79 12 0.0185 1.27 2.98e- 1 2 0.00437 0.300 0.743 24
## 2 A2 2.46 12 0.477 37.4 1.43e-12 2 0.00747 0.585 0.565 24
## 3 A3 2.17 12 0.0840 2.56 2.39e- 2 2 0.0507 1.55 0.233 24
## 4 A4 2.52 12 0.0254 0.858 5.96e- 1 2 0.0179 0.603 0.555 24
## # ... with 4 more variables: MSE <dbl>, CV <dbl>, h2 <dbl>, AS <dbl>
## ---------------------------------------------------------------------------
##
##
##
## Variable ALT_ESP
## ---------------------------------------------------------------------------
## Within-environment ANOVA results
## ---------------------------------------------------------------------------
## # A tibble: 4 x 15
## ENV MEAN DFG MSG FCG PFG DFB MSB FCB PFB DFE
## <chr> <dbl> <int> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <int>
## 1 A1 1.58 12 0.0256 2.03 6.74e- 2 2 0.00728 0.578 0.569 24
## 2 A2 1.31 12 0.363 45.6 1.53e-13 2 0.0180 2.26 0.126 24
## 3 A3 1.08 12 0.0488 1.44 2.14e- 1 2 0.00892 0.264 0.770 24
## 4 A4 1.41 12 0.00919 0.321 9.78e- 1 2 0.0229 0.802 0.460 24
## # ... with 4 more variables: MSE <dbl>, CV <dbl>, h2 <dbl>, AS <dbl>
## ---------------------------------------------------------------------------
##
##
##
## Variable COMPES
## ---------------------------------------------------------------------------
## Within-environment ANOVA results
## ---------------------------------------------------------------------------
## # A tibble: 4 x 15
## ENV MEAN DFG MSG FCG PFG DFB MSB FCB PFB DFE MSE
## <chr> <dbl> <int> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <int> <dbl>
## 1 A1 15.6 12 1.03 0.623 0.802 2 0.363 0.220 0.804 24 1.65
## 2 A2 15.2 12 4.35 5.92 0.000110 2 0.455 0.619 0.547 24 0.734
## 3 A3 14.7 12 1.13 1.14 0.373 2 0.637 0.648 0.532 24 0.984
## 4 A4 15.1 12 3.39 3.50 0.00431 2 0.409 0.422 0.660 24 0.969
## # ... with 3 more variables: CV <dbl>, h2 <dbl>, AS <dbl>
## ---------------------------------------------------------------------------
##
##
##
## Variable DIAMES
## ---------------------------------------------------------------------------
## Within-environment ANOVA results
## ---------------------------------------------------------------------------
## # A tibble: 4 x 15
## ENV MEAN DFG MSG FCG PFG DFB MSB FCB PFB DFE MSE
## <chr> <dbl> <int> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <int> <dbl>
## 1 A1 51.6 12 7.10 3.88 0.00228 2 0.141 0.0772 0.926 24 1.83
## 2 A2 48.7 12 19.7 11.6 0.000000317 2 2.04 1.20 0.319 24 1.70
## 3 A3 47.9 12 18.5 7.63 0.0000138 2 5.19 2.13 0.140 24 2.43
## 4 A4 49.9 12 5.61 1.27 0.297 2 2.03 0.460 0.637 24 4.42
## # ... with 3 more variables: CV <dbl>, h2 <dbl>, AS <dbl>
## ---------------------------------------------------------------------------
##
##
##
## Variable COMP_SAB
## ---------------------------------------------------------------------------
## Within-environment ANOVA results
## ---------------------------------------------------------------------------
## # A tibble: 4 x 15
## ENV MEAN DFG MSG FCG PFG DFB MSB FCB PFB DFE
## <chr> <dbl> <int> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <int>
## 1 A1 29.7 12 11.3 5.51 1.92e-4 2 2.72 1.32 0.285 24
## 2 A2 28.5 12 18.1 17.4 5.47e-9 2 0.00937 0.00898 0.991 24
## 3 A3 28.4 12 14.2 10.1 1.18e-6 2 8.06 5.70 0.00945 24
## 4 A4 29.4 12 5.75 2.74 1.73e-2 2 0.861 0.410 0.668 24
## # ... with 4 more variables: MSE <dbl>, CV <dbl>, h2 <dbl>, AS <dbl>
## ---------------------------------------------------------------------------
##
##
##
## Variable DIAM_SAB
## ---------------------------------------------------------------------------
## Within-environment ANOVA results
## ---------------------------------------------------------------------------
## # A tibble: 4 x 15
## ENV MEAN DFG MSG FCG PFG DFB MSB FCB PFB DFE MSE
## <chr> <dbl> <int> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <int> <dbl>
## 1 A1 16.4 12 1.38 1.17 0.355 2 0.0558 0.0476 0.954 24 1.17
## 2 A2 15.9 12 4.20 5.68 0.000153 2 0.228 0.308 0.738 24 0.739
## 3 A3 15.8 12 1.35 2.13 0.0550 2 1.27 2.01 0.156 24 0.634
## 4 A4 15.8 12 2.49 2.33 0.0372 2 0.318 0.299 0.745 24 1.06
## # ... with 3 more variables: CV <dbl>, h2 <dbl>, AS <dbl>
## ---------------------------------------------------------------------------
##
##
##
## Variable MGE
## ---------------------------------------------------------------------------
## Within-environment ANOVA results
## ---------------------------------------------------------------------------
## # A tibble: 4 x 15
## ENV MEAN DFG MSG FCG PFG DFB MSB FCB PFB DFE MSE
## <chr> <dbl> <int> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <int> <dbl>
## 1 A1 199. 12 597. 2.01 7.01e-2 2 49.8 0.168 0.846 24 297.
## 2 A2 168. 12 3770. 14.9 2.58e-8 2 46.3 0.183 0.834 24 253.
## 3 A3 147. 12 823. 2.94 1.19e-2 2 620. 2.21 0.131 24 280.
## 4 A4 177. 12 836. 1.17 3.59e-1 2 57.6 0.0803 0.923 24 717.
## # ... with 3 more variables: CV <dbl>, h2 <dbl>, AS <dbl>
## ---------------------------------------------------------------------------
##
##
##
## Variable NFIL
## ---------------------------------------------------------------------------
## Within-environment ANOVA results
## ---------------------------------------------------------------------------
## # A tibble: 4 x 15
## ENV MEAN DFG MSG FCG PFG DFB MSB FCB PFB DFE MSE
## <chr> <dbl> <int> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <int> <dbl>
## 1 A1 16.9 12 6.34 2.17 0.0515 2 0.529 0.181 0.836 24 2.92
## 2 A2 15.8 12 4.35 4.63 0.000698 2 2.10 2.23 0.130 24 0.941
## 3 A3 15.8 12 4.81 3.79 0.00267 2 0.640 0.503 0.611 24 1.27
## 4 A4 16.0 12 2.57 1.78 0.111 2 1.20 0.831 0.448 24 1.44
## # ... with 3 more variables: CV <dbl>, h2 <dbl>, AS <dbl>
## ---------------------------------------------------------------------------
##
##
##
## Variable MMG
## ---------------------------------------------------------------------------
## Within-environment ANOVA results
## ---------------------------------------------------------------------------
## # A tibble: 4 x 15
## ENV MEAN DFG MSG FCG PFG DFB MSB FCB PFB DFE MSE
## <chr> <dbl> <int> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <int> <dbl>
## 1 A1 360. 12 2553. 2.52 2.62e-2 2 59.5 0.0587 0.943 24 1015.
## 2 A2 334. 12 9498. 14.1 4.55e-8 2 581. 0.863 0.435 24 674.
## 3 A3 318. 12 3541. 3.48 4.53e-3 2 1172. 1.15 0.333 24 1018.
## 4 A4 343. 12 1842. 1.90 8.67e-2 2 2622. 2.71 0.0868 24 967.
## # ... with 3 more variables: CV <dbl>, h2 <dbl>, AS <dbl>
## ---------------------------------------------------------------------------
##
##
##
## Variable NGE
## ---------------------------------------------------------------------------
## Within-environment ANOVA results
## ---------------------------------------------------------------------------
## # A tibble: 4 x 15
## ENV MEAN DFG MSG FCG PFG DFB MSB FCB PFB DFE MSE
## <chr> <dbl> <int> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <int> <dbl>
## 1 A1 558. 12 5238. 1.43 0.220 2 897. 0.245 0.785 24 3664.
## 2 A2 505. 12 7062. 3.51 0.00430 2 2119. 1.05 0.365 24 2014.
## 3 A3 468. 12 8346. 3.48 0.00451 2 1416. 0.590 0.562 24 2399.
## 4 A4 516. 12 7430. 1.62 0.153 2 3661. 0.797 0.462 24 4595.
## # ... with 3 more variables: CV <dbl>, h2 <dbl>, AS <dbl>
## ---------------------------------------------------------------------------
# Obter dados de todas as variáveis (Coeficiente de variação)
gmd(ind_an, "CV") %>% print_tbl()
## Class of the model: anova_ind
## Variable extracted: CV
| ENV |
ALT_PLANT |
ALT_ESP |
COMPES |
DIAMES |
COMP_SAB |
DIAM_SAB |
MGE |
NFIL |
MMG |
NGE |
| A1 |
4.321 |
7.119 |
8.224 |
2.620 |
4.816 |
6.605 |
8.639 |
10.124 |
8.840 |
10.850 |
| A2 |
4.589 |
6.796 |
5.624 |
2.678 |
3.589 |
5.412 |
9.436 |
6.141 |
7.775 |
8.893 |
| A3 |
8.353 |
17.052 |
6.762 |
3.256 |
4.181 |
5.047 |
11.406 |
7.143 |
10.044 |
10.468 |
| A4 |
6.836 |
12.038 |
6.505 |
4.209 |
4.929 |
6.520 |
15.126 |
7.497 |
9.072 |
13.133 |
# F-máximo
gmd(ind_an, what = "FMAX") %>% print_tbl()
## Class of the model: anova_ind
## Variable extracted: FMAX
| TRAIT |
F_RATIO |
| ALT_PLANT |
2.565 |
| ALT_ESP |
4.243 |
| COMPES |
2.249 |
| DIAMES |
2.593 |
| COMP_SAB |
2.014 |
| DIAM_SAB |
1.851 |
| MGE |
2.840 |
| NFIL |
3.109 |
| MMG |
1.512 |
| NGE |
2.282 |
Anova individual - gafem()
ind_an2 <- gafem(df,
gen = GEN,
rep = BLOCO,
resp = everything(),
by = ENV,
verbose = FALSE)
# Obter dados de todas as variáveis
# P-value
pval <- gmd(ind_an2, what = "Pr(>F)", verbose = FALSE)
print_tbl(pval)
| ENV |
Source |
ALT_PLANT |
ALT_ESP |
COMPES |
DIAMES |
COMP_SAB |
DIAM_SAB |
MGE |
NFIL |
MMG |
NGE |
| A1 |
REP |
0.743 |
0.569 |
0.804 |
0.926 |
0.285 |
0.954 |
0.846 |
0.836 |
0.943 |
0.785 |
| A1 |
GEN |
0.298 |
0.067 |
0.802 |
0.002 |
0.000 |
0.355 |
0.070 |
0.052 |
0.026 |
0.220 |
| A2 |
REP |
0.565 |
0.126 |
0.547 |
0.319 |
0.991 |
0.738 |
0.834 |
0.130 |
0.435 |
0.365 |
| A2 |
GEN |
0.000 |
0.000 |
0.000 |
0.000 |
0.000 |
0.000 |
0.000 |
0.001 |
0.000 |
0.004 |
| A3 |
REP |
0.233 |
0.770 |
0.532 |
0.140 |
0.009 |
0.156 |
0.131 |
0.611 |
0.333 |
0.562 |
| A3 |
GEN |
0.024 |
0.214 |
0.373 |
0.000 |
0.000 |
0.055 |
0.012 |
0.003 |
0.005 |
0.005 |
| A4 |
REP |
0.555 |
0.460 |
0.660 |
0.637 |
0.668 |
0.745 |
0.923 |
0.448 |
0.087 |
0.462 |
| A4 |
GEN |
0.596 |
0.978 |
0.004 |
0.297 |
0.017 |
0.037 |
0.359 |
0.111 |
0.087 |
0.153 |
# Comparação de médias (MGE dentro do ambiente 2)
model_mge_a2 <- ind_an2[[2]][[2]][["MGE"]][["model"]]
pairwise_means <- tukey_hsd(model_mge_a2, "GEN")
print_tbl(pairwise_means)
| term |
group1 |
group2 |
estimate |
conf.low |
conf.high |
p.adj |
sign |
| GEN |
H1 |
H10 |
-28.304 |
-75.822 |
19.214 |
0.612 |
ns |
| GEN |
H1 |
H11 |
-24.589 |
-72.107 |
22.929 |
0.783 |
ns |
| GEN |
H1 |
H12 |
-56.922 |
-104.440 |
-9.404 |
0.010 |
** |
| GEN |
H1 |
H13 |
-19.127 |
-66.645 |
28.391 |
0.949 |
ns |
| GEN |
H1 |
H2 |
30.659 |
-16.859 |
78.177 |
0.498 |
ns |
| GEN |
H1 |
H3 |
2.746 |
-44.772 |
50.263 |
1.000 |
ns |
| GEN |
H1 |
H4 |
9.267 |
-38.251 |
56.785 |
1.000 |
ns |
| GEN |
H1 |
H5 |
-1.955 |
-49.473 |
45.563 |
1.000 |
ns |
| GEN |
H1 |
H6 |
26.832 |
-20.686 |
74.350 |
0.683 |
ns |
| GEN |
H1 |
H7 |
-44.395 |
-91.913 |
3.123 |
0.083 |
ns |
| GEN |
H1 |
H8 |
-75.235 |
-122.753 |
-27.717 |
0.000 |
*** |
| GEN |
H1 |
H9 |
-75.867 |
-123.385 |
-28.349 |
0.000 |
*** |
| GEN |
H10 |
H11 |
3.715 |
-43.803 |
51.233 |
1.000 |
ns |
| GEN |
H10 |
H12 |
-28.618 |
-76.136 |
18.900 |
0.597 |
ns |
| GEN |
H10 |
H13 |
9.177 |
-38.341 |
56.695 |
1.000 |
ns |
| GEN |
H10 |
H2 |
58.963 |
11.445 |
106.481 |
0.007 |
** |
| GEN |
H10 |
H3 |
31.049 |
-16.469 |
78.567 |
0.480 |
ns |
| GEN |
H10 |
H4 |
37.571 |
-9.947 |
85.089 |
0.225 |
ns |
| GEN |
H10 |
H5 |
26.349 |
-21.169 |
73.867 |
0.705 |
ns |
| GEN |
H10 |
H6 |
55.136 |
7.618 |
102.654 |
0.013 |
* |
| GEN |
H10 |
H7 |
-16.091 |
-63.609 |
31.426 |
0.986 |
ns |
| GEN |
H10 |
H8 |
-46.931 |
-94.449 |
0.587 |
0.055 |
ns |
| GEN |
H10 |
H9 |
-47.563 |
-95.081 |
-0.045 |
0.050 |
* |
| GEN |
H11 |
H12 |
-32.333 |
-79.851 |
15.185 |
0.421 |
ns |
| GEN |
H11 |
H13 |
5.462 |
-42.056 |
52.980 |
1.000 |
ns |
| GEN |
H11 |
H2 |
55.248 |
7.730 |
102.766 |
0.013 |
* |
| GEN |
H11 |
H3 |
27.334 |
-20.184 |
74.852 |
0.659 |
ns |
| GEN |
H11 |
H4 |
33.856 |
-13.662 |
81.374 |
0.356 |
ns |
| GEN |
H11 |
H5 |
22.634 |
-24.884 |
70.152 |
0.857 |
ns |
| GEN |
H11 |
H6 |
51.420 |
3.903 |
98.938 |
0.026 |
* |
| GEN |
H11 |
H7 |
-19.807 |
-67.325 |
27.711 |
0.935 |
ns |
| GEN |
H11 |
H8 |
-50.646 |
-98.164 |
-3.129 |
0.029 |
* |
| GEN |
H11 |
H9 |
-51.278 |
-98.796 |
-3.760 |
0.026 |
* |
| GEN |
H12 |
H13 |
37.795 |
-9.723 |
85.313 |
0.218 |
ns |
| GEN |
H12 |
H2 |
87.581 |
40.063 |
135.099 |
0.000 |
**** |
| GEN |
H12 |
H3 |
59.667 |
12.149 |
107.185 |
0.006 |
** |
| GEN |
H12 |
H4 |
66.189 |
18.671 |
113.707 |
0.002 |
** |
| GEN |
H12 |
H5 |
54.967 |
7.449 |
102.485 |
0.014 |
* |
| GEN |
H12 |
H6 |
83.754 |
36.236 |
131.272 |
0.000 |
**** |
| GEN |
H12 |
H7 |
12.526 |
-34.992 |
60.044 |
0.998 |
ns |
| GEN |
H12 |
H8 |
-18.313 |
-65.831 |
29.205 |
0.962 |
ns |
| GEN |
H12 |
H9 |
-18.945 |
-66.463 |
28.573 |
0.952 |
ns |
| GEN |
H13 |
H2 |
49.786 |
2.268 |
97.304 |
0.034 |
* |
| GEN |
H13 |
H3 |
21.872 |
-25.646 |
69.390 |
0.882 |
ns |
| GEN |
H13 |
H4 |
28.394 |
-19.124 |
75.912 |
0.608 |
ns |
| GEN |
H13 |
H5 |
17.172 |
-30.346 |
64.690 |
0.976 |
ns |
| GEN |
H13 |
H6 |
45.959 |
-1.559 |
93.477 |
0.065 |
ns |
| GEN |
H13 |
H7 |
-25.269 |
-72.786 |
22.249 |
0.754 |
ns |
| GEN |
H13 |
H8 |
-56.108 |
-103.626 |
-8.590 |
0.011 |
* |
| GEN |
H13 |
H9 |
-56.740 |
-104.258 |
-9.222 |
0.010 |
* |
| GEN |
H2 |
H3 |
-27.914 |
-75.432 |
19.604 |
0.631 |
ns |
| GEN |
H2 |
H4 |
-21.392 |
-68.910 |
26.126 |
0.896 |
ns |
| GEN |
H2 |
H5 |
-32.614 |
-80.132 |
14.904 |
0.409 |
ns |
| GEN |
H2 |
H6 |
-3.827 |
-51.345 |
43.691 |
1.000 |
ns |
| GEN |
H2 |
H7 |
-75.055 |
-122.573 |
-27.537 |
0.000 |
*** |
| GEN |
H2 |
H8 |
-105.894 |
-153.412 |
-58.376 |
0.000 |
**** |
| GEN |
H2 |
H9 |
-106.526 |
-154.044 |
-59.008 |
0.000 |
**** |
| GEN |
H3 |
H4 |
6.522 |
-40.996 |
54.040 |
1.000 |
ns |
| GEN |
H3 |
H5 |
-4.700 |
-52.218 |
42.818 |
1.000 |
ns |
| GEN |
H3 |
H6 |
24.086 |
-23.432 |
71.604 |
0.804 |
ns |
| GEN |
H3 |
H7 |
-47.141 |
-94.659 |
0.377 |
0.053 |
ns |
| GEN |
H3 |
H8 |
-77.981 |
-125.499 |
-30.463 |
0.000 |
*** |
| GEN |
H3 |
H9 |
-78.612 |
-126.130 |
-31.094 |
0.000 |
*** |
| GEN |
H4 |
H5 |
-11.222 |
-58.740 |
36.296 |
0.999 |
ns |
| GEN |
H4 |
H6 |
17.565 |
-29.953 |
65.082 |
0.972 |
ns |
| GEN |
H4 |
H7 |
-53.663 |
-101.181 |
-6.145 |
0.017 |
* |
| GEN |
H4 |
H8 |
-84.502 |
-132.020 |
-36.985 |
0.000 |
**** |
| GEN |
H4 |
H9 |
-85.134 |
-132.652 |
-37.616 |
0.000 |
**** |
| GEN |
H5 |
H6 |
28.786 |
-18.731 |
76.304 |
0.589 |
ns |
| GEN |
H5 |
H7 |
-42.441 |
-89.959 |
5.077 |
0.112 |
ns |
| GEN |
H5 |
H8 |
-73.281 |
-120.798 |
-25.763 |
0.000 |
*** |
| GEN |
H5 |
H9 |
-73.912 |
-121.430 |
-26.394 |
0.000 |
*** |
| GEN |
H6 |
H7 |
-71.227 |
-118.745 |
-23.709 |
0.001 |
*** |
| GEN |
H6 |
H8 |
-102.067 |
-149.585 |
-54.549 |
0.000 |
**** |
| GEN |
H6 |
H9 |
-102.698 |
-150.216 |
-55.180 |
0.000 |
**** |
| GEN |
H7 |
H8 |
-30.840 |
-78.358 |
16.678 |
0.490 |
ns |
| GEN |
H7 |
H9 |
-31.471 |
-78.989 |
16.047 |
0.460 |
ns |
| GEN |
H8 |
H9 |
-0.631 |
-48.149 |
46.887 |
1.000 |
ns |
# comparações de médias com o pacote emmeans
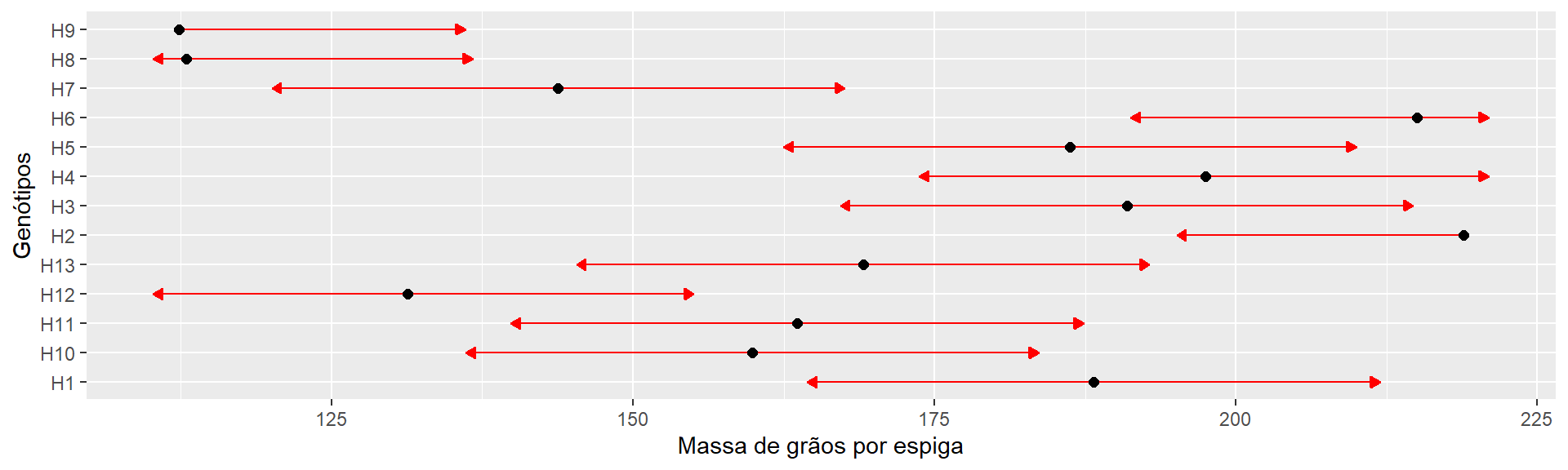
(means <- emmeans(model_mge_a2, "GEN"))
## GEN emmean SE df lower.CL upper.CL
## H1 188 9.18 24 169.3 207
## H10 160 9.18 24 141.0 179
## H11 164 9.18 24 144.7 183
## H12 131 9.18 24 112.3 150
## H13 169 9.18 24 150.1 188
## H2 219 9.18 24 199.9 238
## H3 191 9.18 24 172.0 210
## H4 197 9.18 24 178.5 216
## H5 186 9.18 24 167.3 205
## H6 215 9.18 24 196.1 234
## H7 144 9.18 24 124.9 163
## H8 113 9.18 24 94.0 132
## H9 112 9.18 24 93.4 131
##
## Results are averaged over the levels of: REP
## Confidence level used: 0.95
plot(means,
comparisons = TRUE,
CIs = FALSE,
xlab = "Massa de grãos por espiga",
ylab = "Genótipos")